Difference between revisions of "Description of GenGIS plugins"
| Line 19: | Line 19: | ||
=Beta Diversity= | =Beta Diversity= | ||
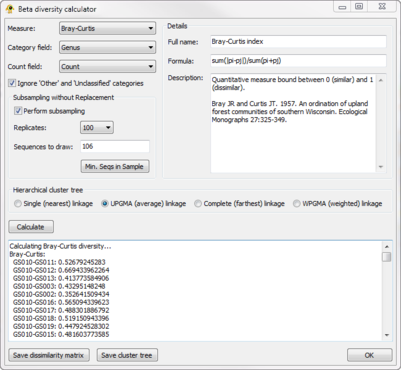
| − | The ''Beta Diversity'' plugin calculates [http://en.wikipedia.org/wiki/Beta_diversity beta diversity] between active locations (Fig. 3). The resulting biotic dissimilarity matrix can be saved to file and visualized in GenGIS using the ''Dissimilarity Matrix Viewer'' plugin. It currently calculate 9 measures of beta diversity (e.g., Bray-Curtis, Jaccard) across any field defined in your ''Sequence File''. Sequences classified as Other or Unclassified can be optionally ignored during the calculation of beta diversity. In order to account for unequal sampling depth, subsampling with replacement (i.e., jackknifing) can be performed and the mean beta-diversity between jackknifed samples reported. Hierarchical cluster trees indicating the relative similarity of locations can be produced and used as an input 'Tree File' to GenGIS. | + | The ''Beta Diversity'' plugin calculates [http://en.wikipedia.org/wiki/Beta_diversity beta diversity] between active locations (Fig. 3). The resulting biotic dissimilarity matrix can be saved to file and visualized in GenGIS using the ''Dissimilarity Matrix Viewer'' plugin. It currently calculate 9 measures of beta diversity (e.g., Bray-Curtis, Jaccard) across any field defined in your ''Sequence File''. Sequences classified as Other or Unclassified can be optionally ignored during the calculation of beta diversity. In order to account for unequal sampling depth, subsampling with replacement (i.e., jackknifing) can be performed and the mean beta-diversity between jackknifed samples reported. Hierarchical cluster trees indicating the relative similarity of locations can be produced and used as an input ''Tree File'' to GenGIS. |
[[Image:BetaDiversityPlugin.png|thumb|center|401px|Figure 3. Beta Diversity Plugin]] | [[Image:BetaDiversityPlugin.png|thumb|center|401px|Figure 3. Beta Diversity Plugin]] | ||
Revision as of 17:56, 24 March 2013
GenGIS provides the following Python plugins which can be accessed through the Plugins menu.
Contents
Alpha Diversity
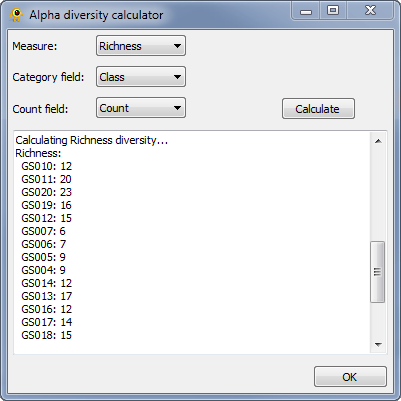
The Alpha Diversity plugin calculates alpha diversity for active locations. It currently calculate richness, Shannon, and Simpson alpha diversity. To calculate alpha diversity, you must select the Measure you wish to calculate and the Category field in your sequence file over which diversity will be calculate (Fig. 1). You may optionally select a Count field which indicates the number of times a given sequence is observed at a location. Pressing Calculate causes alpha diversity to be calculated. Results are reported within the plugin and added to the location table for use within GenGIS and other plugins.
Alpha Diversity Visualizer
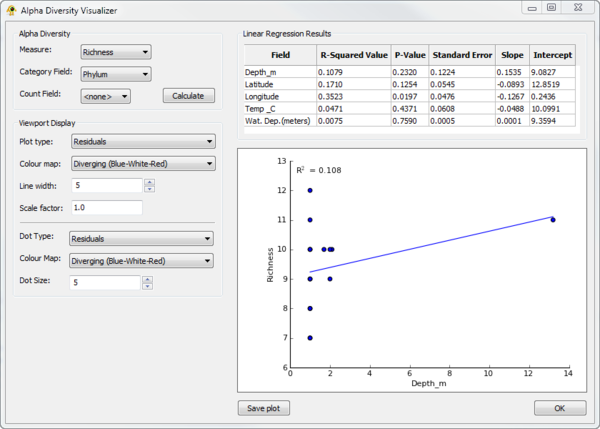
The Alpha Diversity Visualizer plugin can calculate alpha diversity for active locations, regress alpha diversity against location specific metadata, and produce visualizations of the resulting linear regression analysis. It currently calculate richness, Shannon, and Simpson alpha diversity. To calculate alpha diversity, you must select the Measure you wish to calculate and the Category field in your sequence file over which diversity will be calculate (Fig. 2). You may optionally select a Count field which indicates the number of times a given sequence is observed at a location. Pressing Calculate causes alpha diversity to be calculated. Linear regression results of alpha diversity versus all numeric fields associated with locations are reported within the Linear Regression Results table. Selecting a row within this table causes a linear regression scatter plot of alpha diversity versus the selected Field to be generated. The Viewport Display section allows different Viewport visualization to be produced.
Bar Graph
Coming soon.
Beta Diversity
The Beta Diversity plugin calculates beta diversity between active locations (Fig. 3). The resulting biotic dissimilarity matrix can be saved to file and visualized in GenGIS using the Dissimilarity Matrix Viewer plugin. It currently calculate 9 measures of beta diversity (e.g., Bray-Curtis, Jaccard) across any field defined in your Sequence File. Sequences classified as Other or Unclassified can be optionally ignored during the calculation of beta diversity. In order to account for unequal sampling depth, subsampling with replacement (i.e., jackknifing) can be performed and the mean beta-diversity between jackknifed samples reported. Hierarchical cluster trees indicating the relative similarity of locations can be produced and used as an input Tree File to GenGIS.
Dissimilarity Matrix Viewer
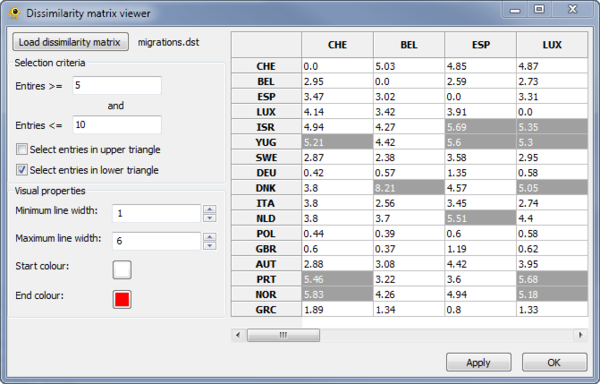
The Dissimilarity Matrix Viewer plugin provides functionality for visualizing a matrix which indicates the dissimilarity between all pairs of locations. The dissimilarity matrix must be in the following format, where a \t indicates a tab:
3 A\t0\t2\t3 B\t1\t0\t4 C\t3\t5\t0
The first line indicates the number of locations and each of the following rows gives the dissimilarity values for the specified location. The location names (first column) must match those in your location file. The upper and lower triangles of the matrix can be different. For example, in this HIV-1 data set, the two triangle indicate import and export rates.
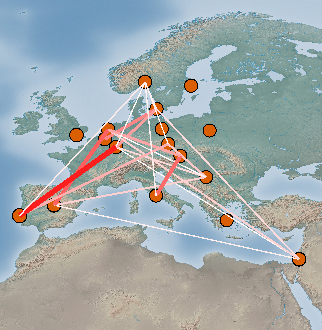
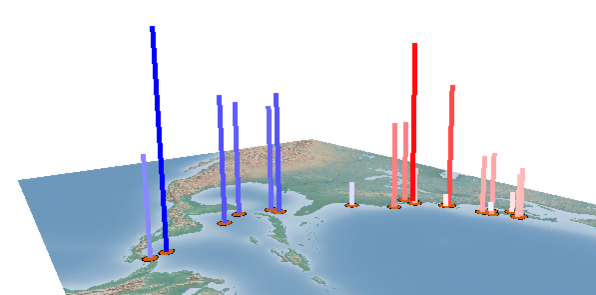
Elements in the matrix are selected by setting the Selection criteria (Fig. 4). Lines between the selected pairs are displayed in the Viewport using the specified Visual properties (Fig. 5). To update the Viewport display click Apply.
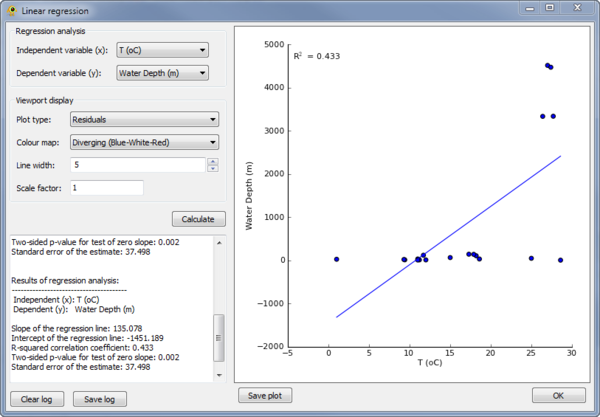
Linear Regression
The Linear Regression plugin can be used to perform a linear regress between any two variables in the Location Table (see Location Table Viewer below). To perform the regression, the independent and dependent variables must be specified in the Regression analysis section of the plugin (Fig. 6). The results of the regression are reported within the plugin and shown as a scatter plot. A visualization within the GenGIS Viewport is also generated based on the properties set in the Viewport display section of the plugin (Fig. 7).
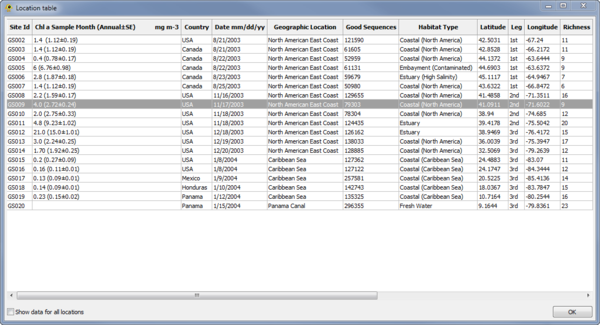
Location Table Viewer
The Location Table Viewer plugin display a table indicating the metadata associated with each location (Fig. 8). Other plugins and custom Python scripts can be used to add data to the Location Table. By default, only data for active locations is shown. To show data for all locations check the Show data for all locations checkbox.
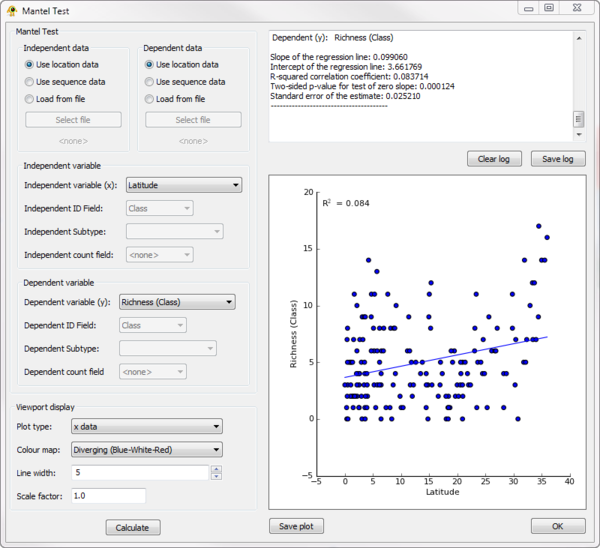
Mantel
- Requirements: R with the ade4 library must be installed on your system (see the GenGIS manual).
The Mantel plugin can be used to perform a Mantel test between any two variables in the Location Table or Sequence Table.
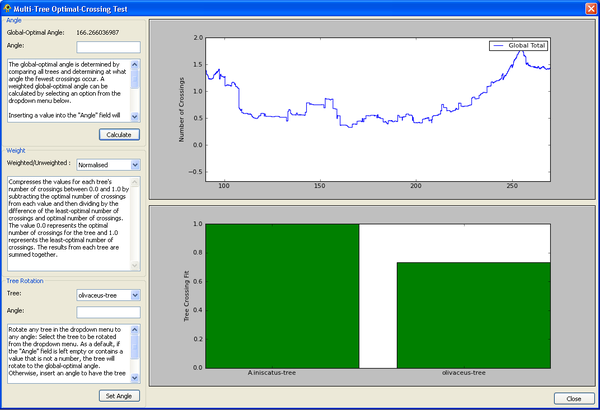
Multi-Tree Optimal-Crossing Test
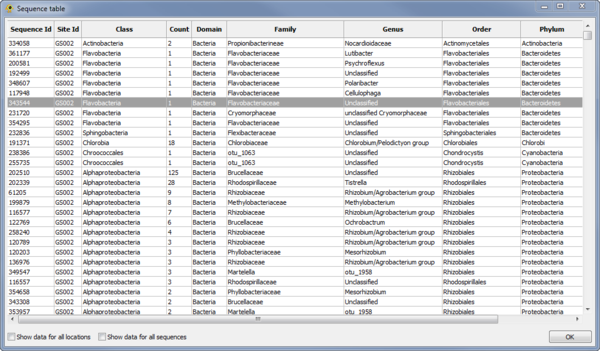
Sequence Table Viewer
The Sequence Table Viewer plugin display a table indicating the metadata associated with each sequence (Fig. 11). Other plugins and custom Python scripts can be used to add data to the Sequence Table. By default, only data for active locations and active sequences is shown. To show data for all locations check the Show data for all locations checkbox. To show data for all sequences check the Show data for all sequences checkbox.