Difference between revisions of "SPR Supertrees example datasets"
From Bioinformatics Software
Jump to navigationJump to search| Line 3: | Line 3: | ||
== 244 Taxa Bacterial Supertree == | == 244 Taxa Bacterial Supertree == | ||
| − | [[Image:244_taxa_supertree.png]] | + | [[Image:244_taxa_supertree.png|thumb|457x540px]] |
| − | |||
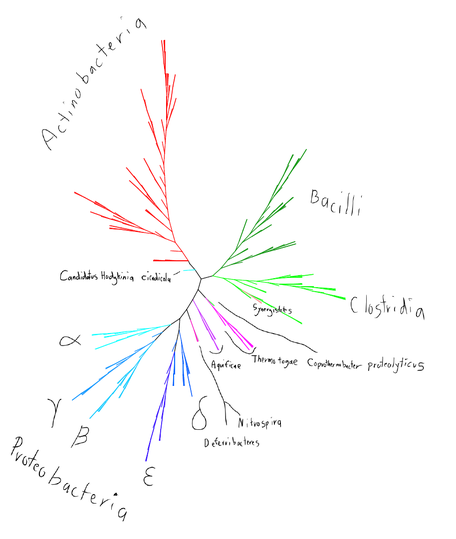
A recent presentation at iEvoBio 2012 demonstrated a 244-taxa bacterial SPR Supertree constructed from a subset of the data from the 159,905 prokaryotic phylogenetic trees from “Telling the Whole Story in a 10,000-Genome World” (PMID:21714939). The full trees are available at Dryad (doi:10.5061/dryad.gf39c8dc). To reconstruct this subset from the full dataset, you can use the -include_only option of spr_supertree with [[media:244_taxa_chosen|this text file of the chosen taxa]] and the parameters -allow_multi -support 0.8 -simple_unrooted. With the -valid_trees option, this will print out the trees and quit. | A recent presentation at iEvoBio 2012 demonstrated a 244-taxa bacterial SPR Supertree constructed from a subset of the data from the 159,905 prokaryotic phylogenetic trees from “Telling the Whole Story in a 10,000-Genome World” (PMID:21714939). The full trees are available at Dryad (doi:10.5061/dryad.gf39c8dc). To reconstruct this subset from the full dataset, you can use the -include_only option of spr_supertree with [[media:244_taxa_chosen|this text file of the chosen taxa]] and the parameters -allow_multi -support 0.8 -simple_unrooted. With the -valid_trees option, this will print out the trees and quit. | ||
The constructed tree is available in [[media:244_taxa_supertree|newick]] format. | The constructed tree is available in [[media:244_taxa_supertree|newick]] format. | ||
Revision as of 07:20, 9 July 2012
244 Taxa Bacterial Supertree
A recent presentation at iEvoBio 2012 demonstrated a 244-taxa bacterial SPR Supertree constructed from a subset of the data from the 159,905 prokaryotic phylogenetic trees from “Telling the Whole Story in a 10,000-Genome World” (PMID:21714939). The full trees are available at Dryad (doi:10.5061/dryad.gf39c8dc). To reconstruct this subset from the full dataset, you can use the -include_only option of spr_supertree with this text file of the chosen taxa and the parameters -allow_multi -support 0.8 -simple_unrooted. With the -valid_trees option, this will print out the trees and quit.
The constructed tree is available in newick format.