Difference between revisions of "Main Page"
From Bioinformatics Software
Jump to navigationJump to search| Line 7: | Line 7: | ||
Key: P = phylogenetics, S = statistics, B = biogeography, V = visualization, G = genomics, M = metagenomics, L = lateral genetic transfer, A = sequence alignment | Key: P = phylogenetics, S = statistics, B = biogeography, V = visualization, G = genomics, M = metagenomics, L = lateral genetic transfer, A = sequence alignment | ||
| − | + | Join our [http://ratite.cs.dal.ca/software_email_list/subscribe mailing list] to keep informed about new developments. | |
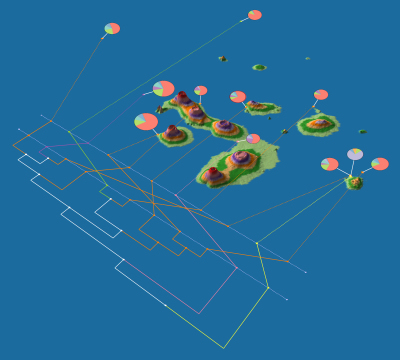
* [http://kiwi.cs.dal.ca/GenGIS GenGIS]: an application that allows users to combine digital map data with information about biological sequences collected from the environment. GenGIS provides a 3D graphical interface in which the user can navigate and explore the data, as well as a Python interface that allows easy scripting of statistical analyses using the Rpy libraries. (PSBVM) | * [http://kiwi.cs.dal.ca/GenGIS GenGIS]: an application that allows users to combine digital map data with information about biological sequences collected from the environment. GenGIS provides a 3D graphical interface in which the user can navigate and explore the data, as well as a Python interface that allows easy scripting of statistical analyses using the Rpy libraries. (PSBVM) | ||
Revision as of 13:32, 20 May 2010
Welcome to the Bioinformatics Software and Resources page.
Software
Key: P = phylogenetics, S = statistics, B = biogeography, V = visualization, G = genomics, M = metagenomics, L = lateral genetic transfer, A = sequence alignment
Join our mailing list to keep informed about new developments.
- GenGIS: an application that allows users to combine digital map data with information about biological sequences collected from the environment. GenGIS provides a 3D graphical interface in which the user can navigate and explore the data, as well as a Python interface that allows easy scripting of statistical analyses using the Rpy libraries. (PSBVM)
- PICA: genotype-phenotype data mining software (G).
- rSPR: software to calculate rooted subtree-prune-and-regraft distances and rooted agreement forests. (PL)
- STAMP: a software package for analyzing metagenomic profiles that promotes ‘best practices’ in choosing appropriate statistical techniques and reporting results. (SMV)
- Radié: a tool that allows characters to be visualized against the background of a phylogenetic tree. The software includes several different visual and numeric representations of the ‘convexity’ of a given character, in other words the extent to which different character traits form distinct groups within the tree. (PV)
- EvolSimulator: a simulation test bed for hypotheses of genome evolution. (PL)
- EEEP (Efficient Evaluation of Edit Paths): software to infer putative pathways of lateral genetic transfer by comparing gene trees against a rooted reference tree (PL)
- WOOF: a tool designed to rigourously apply the principle of visual alignment validation. (SA)
- GANN: a machine learning method designed with the complexities of transcriptional regulation in mind.
- VAREB [to be added]
Web Services
- SeqMonitor
- MOAMap
- MOA (to be added)
- Visual MOA (to be added)
- MANUEL
Datasets
- Datasets used in GenGIS (Parks et al., Genome Research 2009)
- STAMP datasets
- Lateral genetic transfer in 144 genomes dataset (Beiko et al., Proc. Natl. Acad. Sci. 2005)
- Simulated data for 'The impact of reticulate evolution on genome phylogeny' (Beiko et al., Systematic Biology 2008) [to be added]
Contributors
- Robert Beiko
- Christian Blouin
- Donovan Parks
- Chris Whidden
- Norman MacDonald