Difference between revisions of "STAMP"
| Line 17: | Line 17: | ||
=== Documentation === | === Documentation === | ||
| − | * [[Quick installation instructions for STAMP|Quick installation instructions]] (Microsoft Windows, Apple's Mac OS X, Command-line interface) | + | * [[Quick installation instructions for STAMP|Quick installation instructions]] (Microsoft Windows, Apple's Mac OS X, Linux, Command-line interface) |
* [[Media:STAMP_Users_Guide.pdf|User's Guide]] | * [[Media:STAMP_Users_Guide.pdf|User's Guide]] | ||
* [[STAMP FAQs|FAQs]] | * [[STAMP FAQs|FAQs]] | ||
Revision as of 15:28, 17 July 2010
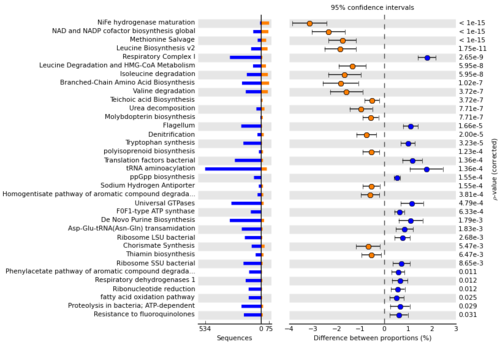
STAMP (Statistical Analysis of Metagenomic Profiles) is a software package for analyzing metagenomic profiles (e.g., a phylogenetic profile indicating the number of marker genes assigned to different taxonomic units or a functional profile indicating the number of sequences assigned to different biological subsystems or pathways) that promotes ‘best practices’ in choosing appropriate statistical techniques and reporting results. It encourages the use of effect sizes and confidence intervals in assessing biological importance. A user friendly, graphical interface permits easy exploration of statistical results and generation of publication quality plots for inferring the biological relevance of features in a metagenomic profile. STAMP is open source, extensible via a plugin framework, and available for all major platforms.
Announcements
- June 16, 2010: STAMP v1.06a released for Mac OS X. The data files required for COG annotation were missing from v1.06.
- May 27, 2010: STAMP v1.06 released. Improves error reporting on incorrect file formats and potential software bugs.
- May 18, 2010: STAMP v1.05 released. Minor bug fixes.
- Previous announcements
Mailing list
- Join our mailing list to keep informed about STAMP developments.
Documentation
- Quick installation instructions (Microsoft Windows, Apple's Mac OS X, Linux, Command-line interface)
- User's Guide
- FAQs
- Version history
Downloads
- STAMP v1.06 executable for Microsoft Windows (~17MB)
- STAMP v1.06a executable for Mac OS X Leopard and Snow Leopard (~72MB) [Beta version: if you experience problems please let us know.]
- STAMP v1.06 source code
Examples
Citing STAMP
If you use STAMP in your research, please cite:
Parks, D.H. and Beiko, R.G. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics, 26, 715-721. (Abstract)
Contact Information
STAMP is in active development and we are interested in discussing all potential applications of this software. We encourage you to send us suggestions for new features. Suggestions, comments, and bug reports can be sent to Rob Beiko (beiko [at] cs.dal.ca). If reporting a bug, please provide as much information as possible and a simplified version of the data set which causes the bug. This will allow us to quickly resolve the issue.
Funding
The development and deployment of STAMP has been supported by several organizations:
- Genome Atlantic
- The Dalhousie Centre for Comparative Genomics and Evolutionary Bioinformatics, and the Tula Foundation
- The Natural Sciences and Engineering Research Council of Canada
- The Dalhousie Faculty of Computer Science