Difference between revisions of "STAMP"
| (83 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
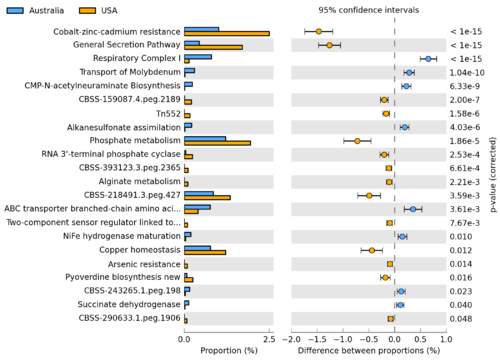
[[Image:STAMP2_EBPR_Aphosphatis_ExtendedErrorBar.png|thumb|right|500px|Using STAMP to identify SEED subsystems which are differentially abundant between ''Candidatus'' Accumulibacter phosphatis sequences obtained from a pair of enhanced biological phosphorus removal (EBPR) sludge metagenomes(data originally described in [http://www.ncbi.nlm.nih.gov/pubmed/20130030 Parks and Beiko, 2010]).]] | [[Image:STAMP2_EBPR_Aphosphatis_ExtendedErrorBar.png|thumb|right|500px|Using STAMP to identify SEED subsystems which are differentially abundant between ''Candidatus'' Accumulibacter phosphatis sequences obtained from a pair of enhanced biological phosphorus removal (EBPR) sludge metagenomes(data originally described in [http://www.ncbi.nlm.nih.gov/pubmed/20130030 Parks and Beiko, 2010]).]] | ||
| − | STAMP | + | STAMP is a software package for analyzing taxonomic or metabolic profiles that promotes ‘best practices’ in choosing appropriate statistical techniques and reporting results. Statistical hypothesis tests for pairs of samples or groups of samples is support along with a wide range of exploratory plots. STAMP encourages the use of effect sizes and confidence intervals in assessing biological importance. A user friendly, graphical interface permits easy exploration of statistical results and generation of publication quality plots for inferring the biological relevance of features in a metagenomic profile. STAMP is open source, extensible via a plugin framework, and available for all major platforms. |
=== Announcements === | === Announcements === | ||
| − | * | + | * June 26, 2015: STAMP v2.1.3 released. Minor bug fix to scatter plot to properly handle profiles contained a single feature. |
| + | * June 15, 2015: STAMP v2.1.2 released. Minor enhancements to extended error bar, heatmap, and profile bar plots. | ||
| + | * June 7, 2015: STAMP v2.1.1 released. Resolves issue with v2.1.0 installation. | ||
| + | * June 4, 2015: STAMP v2.1.0 released. Resolves the _hierarchy_wrap issue. Requires numpy >= 1.9.1, scipy >= 0.15.1, matplotlib >= 1.4.2. | ||
* [[Previous announcements]] | * [[Previous announcements]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
=== Documentation === | === Documentation === | ||
| − | * [[Quick installation instructions for STAMP|Quick installation instructions]] (Microsoft Windows, Apple's Mac OS X | + | * [[Quick installation instructions for STAMP|Quick installation instructions]] (Microsoft Windows, Linux, Apple's Mac OS X) |
| − | * [[Media: | + | * [[Media:STAMP_Users_Guide.zip|User's Guide]] |
| + | * [https://groups.google.com/forum/?hl=en#!forum/stamp_help Google Group] | ||
* [[STAMP FAQs|FAQs]] | * [[STAMP FAQs|FAQs]] | ||
* [[STAMP version history|Version history]] | * [[STAMP version history|Version history]] | ||
| Line 23: | Line 23: | ||
''Please uninstall previous versions of STAMP before installing a new release.'' | ''Please uninstall previous versions of STAMP before installing a new release.'' | ||
| − | * [ | + | * [https://github.com/dparks1134/STAMP/releases/download/v2.1.3/STAMP_2_1_3.exe STAMP v2.1.3] setup package for Microsoft Windows (~42MB) |
| − | * | + | * Linux and OS X users can follow the instructions to [[Quick installation instructions for STAMP|install from source]] |
| − | * [ | + | * [https://github.com/dparks1134/STAMP STAMP GitHub Repository] |
* [[Previous versions|Previous versions]] | * [[Previous versions|Previous versions]] | ||
| + | |||
| + | === Additional Scripts === | ||
| + | |||
| + | * [[Media:CheckHierarchy.zip|checkHierarchy.py]]: identify entries within a STAMP profile that are not strictly hierarchical | ||
=== Examples === | === Examples === | ||
| Line 34: | Line 38: | ||
== Citing STAMP == | == Citing STAMP == | ||
| − | If you use STAMP in your research, | + | If you use STAMP v2+ in your research, we appreciate you citing: |
| + | |||
| + | * Parks DH, Tyson GW, Hugenholtz P, Beiko RG. (2014). [http://bioinformatics.oxfordjournals.org/cgi/content/abstract/btu494?ijkey=T0tIsUilyLV0x3w&keytype=ref STAMP: Statistical analysis of taxonomic and functional profiles]. ''Bioinformatics'', 30, 3123-3124. | ||
| + | |||
| + | |||
| + | STAMP v1 manuscript: | ||
| − | + | * Parks DH and Beiko RG. (2010). [http://bioinformatics.oxfordjournals.org/cgi/content/short/26/6/715 Identifying biologically relevant differences between metagenomic communities]. ''Bioinformatics'', 26, 715-721. | |
== Contact Information == | == Contact Information == | ||
| − | STAMP is in active development and we are interested in discussing all potential applications of this software. We | + | STAMP is in active development and we are interested in discussing all potential applications of this software. We recommend you send suggestions, comments, and bug reports to the [[https://groups.google.com/forum/?hl=en#!forum/stamp_help STAMP Google Group]] so all commuity members can participate and benefit from the discussions. If reporting a bug, please provide as much information as possible and a simplified version of the data set which causes the bug. This will allow us to quickly resolve the issue. If you need to contact the authors of STAMP privately, please email Rob Beiko (beiko [at] cs.dal.ca). |
== Funding == | == Funding == | ||
Latest revision as of 04:44, 26 June 2015
STAMP is a software package for analyzing taxonomic or metabolic profiles that promotes ‘best practices’ in choosing appropriate statistical techniques and reporting results. Statistical hypothesis tests for pairs of samples or groups of samples is support along with a wide range of exploratory plots. STAMP encourages the use of effect sizes and confidence intervals in assessing biological importance. A user friendly, graphical interface permits easy exploration of statistical results and generation of publication quality plots for inferring the biological relevance of features in a metagenomic profile. STAMP is open source, extensible via a plugin framework, and available for all major platforms.
Announcements
- June 26, 2015: STAMP v2.1.3 released. Minor bug fix to scatter plot to properly handle profiles contained a single feature.
- June 15, 2015: STAMP v2.1.2 released. Minor enhancements to extended error bar, heatmap, and profile bar plots.
- June 7, 2015: STAMP v2.1.1 released. Resolves issue with v2.1.0 installation.
- June 4, 2015: STAMP v2.1.0 released. Resolves the _hierarchy_wrap issue. Requires numpy >= 1.9.1, scipy >= 0.15.1, matplotlib >= 1.4.2.
- Previous announcements
Documentation
- Quick installation instructions (Microsoft Windows, Linux, Apple's Mac OS X)
- User's Guide
- Google Group
- FAQs
- Version history
Downloads
Please uninstall previous versions of STAMP before installing a new release.
- STAMP v2.1.3 setup package for Microsoft Windows (~42MB)
- Linux and OS X users can follow the instructions to install from source
- STAMP GitHub Repository
- Previous versions
Additional Scripts
- checkHierarchy.py: identify entries within a STAMP profile that are not strictly hierarchical
Examples
Citing STAMP
If you use STAMP v2+ in your research, we appreciate you citing:
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics, 30, 3123-3124.
STAMP v1 manuscript:
- Parks DH and Beiko RG. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics, 26, 715-721.
Contact Information
STAMP is in active development and we are interested in discussing all potential applications of this software. We recommend you send suggestions, comments, and bug reports to the [STAMP Google Group] so all commuity members can participate and benefit from the discussions. If reporting a bug, please provide as much information as possible and a simplified version of the data set which causes the bug. This will allow us to quickly resolve the issue. If you need to contact the authors of STAMP privately, please email Rob Beiko (beiko [at] cs.dal.ca).
Funding
The development and deployment of STAMP has been supported by several organizations:
- Genome Atlantic
- The Dalhousie Centre for Comparative Genomics and Evolutionary Bioinformatics, and the Tula Foundation
- Killam Trust
- The Natural Sciences and Engineering Research Council of Canada
- The Dalhousie Faculty of Computer Science