Difference between revisions of "Main Page"
| Line 34: | Line 34: | ||
Please cite GenGIS if you use it in your work. Currently the best citation for GenGIS is: | Please cite GenGIS if you use it in your work. Currently the best citation for GenGIS is: | ||
| − | '''Parks, D.H., Porter, M., Churcher, S., Wang, S., Blouin, C., Whalley, J., Brooks, S. and Beiko, R.G. (2009). GenGIS: A geospatial information system for genomic data. ''Genome Research, 19: 1896-1904. | + | '''Parks, D.H., Porter, M., Churcher, S., Wang, S., Blouin, C., Whalley, J., Brooks, S. and Beiko, R.G. (2009). GenGIS: A geospatial information system for genomic data. ''Genome Research'', 19: 1896-1904. ([http://genome.cshlp.org/content/19/10/1896.abstract?sid=5fcec469-a13c-44f9-9f40-9e913e1c0ddf Abstract])''' |
Revision as of 07:32, 24 August 2011
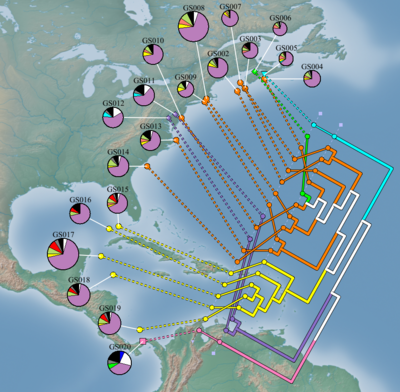
GenGIS (pronounced 'Genghis' like Genghis Khan) is a bioinformatics application that allows users to combine digital map data with information about biological sequences collected from the environment. GenGIS provides a 3D graphical interface in which the user can navigate and explore the data, as well as a Python interface that allows easy scripting of statistical analyses using the Rpy libraries.
Below you can find information about the software, data sources, tutorials, and the manual. The manual is still under development, but the tutorials give a good overview of how to use the software.
Getting Started
- Join our mailing list to keep informed about GenGIS developments
- Download MapMaker (provides easy access to maps compatible with GenGIS)
- Visit the documentation page (Tutorials, manual, FAQ's)
Announcements
- (Sept. 14, 2010) GenGIS v1.08 release for Windows and OS X. A list of new features and bug fixes can be found here.
- (July 6, 2010) GenGIS places 2nd in the iEvoBio visualisation challenge.
Examples
We are building a gallery of example applications using GenGIS, including tracking of the current H1N1 outbreak and the spread of HIV-1 subtype B in Europe.
Citing GenGIS
Please cite GenGIS if you use it in your work. Currently the best citation for GenGIS is:
Parks, D.H., Porter, M., Churcher, S., Wang, S., Blouin, C., Whalley, J., Brooks, S. and Beiko, R.G. (2009). GenGIS: A geospatial information system for genomic data. Genome Research, 19: 1896-1904. (Abstract)
Other papers and conference proceedings related to GenGIS:
- Parks, D.H., MacDonald, N.J., and Beiko, R.G. (2009). Tracking the evolution and geographic spread of Influenza A. PLoS Currents: Influenza August 27:RRN1014. (Article)
- Parks, D.H. and Beiko, R.G. (2009). Quantitative visualizations of hierarchically organized data in a geographic context. Geoinformatics 2009, Fairfax, VA. Donovan won the best student paper award for his presentation. (Abstract)
- Beiko, R., Whalley, J., Wang, S., Clair, H., Smolyn, G., Churcher, S., Porter, M., Blouin, C., & Brooks, S. (2008). Spatial analysis and visualization of genetic biodiversity. Free and Open Source Software for Geospatial (FOSS4G), Cape Town, South Africa (September-October 2008). (Abstract) (Full Paper)
- Wang, S., Beiko, R.G., & Brooks, S. (2007). Collapsible 3D Terrains for GIS Visualization. Geovisualization, NUI Maynooth (September 2007). (Full Paper)
Contact Information
GenGIS is in active development and we are interested in discussing all potential applications of this software. We encourage you to send us suggestions for new features. Suggestions, comments, and bug reports can be sent to Rob Beiko (beiko [at] cs.dal.ca). If reporting a bug, please provide as much information as possible and a simplified version of the data set which causes the bug. This will allow us to quickly resolve the issue.
Funding of GenGIS
The development and deployment of GenGIS has been supported by several organizations:
- Genome Atlantic
- The Dalhousie Centre for Comparative Genomics and Evolutionary Bioinformatics, and the Tula Foundation
- The Natural Sciences and Engineering Research Council of Canada
- The Dalhousie Faculty of Computer Science