Difference between revisions of "GenGIS Tutorials"
| Line 17: | Line 17: | ||
* [[Media:GenGIS_Katydids_Data.zip|Download katydid data set]] | * [[Media:GenGIS_Katydids_Data.zip|Download katydid data set]] | ||
| + | |||
=== Tutorial 2: Marine Microbial Communities from the Atlantic Seaboard === | === Tutorial 2: Marine Microbial Communities from the Atlantic Seaboard === | ||
Revision as of 01:03, 26 August 2009
We have put together a few tutorials to get you quickly started with GenGIS.
Contents
Tutorial 0: Construct a Custom Digital Elevation Maps (DEM)
Instructions on how to contruct a custom DEM using the data available from the GTOPO30 site is available here.
Tutorial 1: Banza Katydids of the Hawaiian Islands
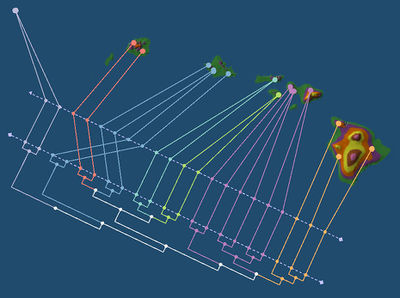
In this tutorial, we consider the phylogenetic tree of Banza katydids (acoustic insects) from the Hawaiian Islands recently recovered by Shapiro et al. [1]. GenGIS will be used to investigate whether or not this phylogeny is related to the geography of the Hawaiian Islands.
Tutorial 2: Marine Microbial Communities from the Atlantic Seaboard
In this tutorial, we examine samples collected as part of the Global Ocean Sampling metagenomic survey [2] in order to investigate the influence of environmental factors on the composition of microbial communities from marine ecosystems.
Tutorial 3: Custom Data Visualizations and Movies
We have put together a number of examples illustrating how custom data visualizations and movies can be created using the API exposed to the Python console:
- Here is a list of all API functions.
- A few short examples of using the API are available in the GenGIS manual.
- An example showing how GenGIS can be used to visualize the mobility rates of HIV-1 subtype B in Europe as proposed by Paraskevis et al. (2009) has been posted here. This example includes a Python class demonstrating how a custom visualization can be made.
- A video showing the spread of the 2009 H1N1 influenza outbreak is available here along with all data and code used to create the video. The Python class demonstrates how the properties of location sites can be accessed and modified using the API.
- A video showing the spread of polymorphic amino acid sites during the 2009 H1N1 outbreak is available here. The included Python class demonstrates how how pie charts can be accessed and modified using the API.
- A video showing the geophylogeny of the 2009 H1N1 outbreak is available here. The included Python class demonstrates how geophylogenies can be accessed and modified using the API.
References
[1] L. H. Shapiro, J. S. Strazanac, and G. K. Roderick. (2006) Molecular phylogeny of Banza (Orthoptera: Tettigoniidae), the endemic katydids of the Hawaiian Archipelago. Mol. Phylogenet. Evol. 41:53-63. Pubmed
[2] D. Rusch, A. Halpern, G. Sutton, et al., (2007) "The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific," PLoS Biol. 5:e77. PubMed