Difference between revisions of "GenGIS Tutorials"
| (46 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
We have put together a few tutorials to get you quickly started with GenGIS. | We have put together a few tutorials to get you quickly started with GenGIS. | ||
| − | == | + | == Short video tutorials == |
| + | These videos show how different tasks can be performed in GenGIS. | ||
| − | + | === Creating a basemap === | |
| + | Here is a short illustration of basemap creation using both raster and vector data. [[Media:Basemap larger x264.mp4 | View Tutorial]] (3:25) | ||
| − | + | === Querying GBIF === | |
| + | From version 2.1.1 you can add data directly from the Global Biodiversity Information Facility (GBIF). Here is a [[Media:GBIF x264.mp4 | tutorial showing the retrieval of cougar and jaguar records from GBIF]]. (11:12) | ||
| + | == Walkthrough tutorials with files == | ||
| + | |||
| + | === Tutorial 0: Obtaining Custom Digital Elevation Maps (DEM) === | ||
| + | |||
| + | The simple way to obtain DEMs suitable for use in GenGIS is described [[Obtaining DEM from ORNL DAAC|here]]. An older set of instructions on how to manually contruct a custom DEM is available [[GTOP030 Tutorial|here]]. You can also obtain 2D maps compatible with GenGIS using [[MapMaker|MapMaker]]. | ||
=== Tutorial 1: ''Banza'' Katydids of the Hawaiian Islands === | === Tutorial 1: ''Banza'' Katydids of the Hawaiian Islands === | ||
| Line 12: | Line 20: | ||
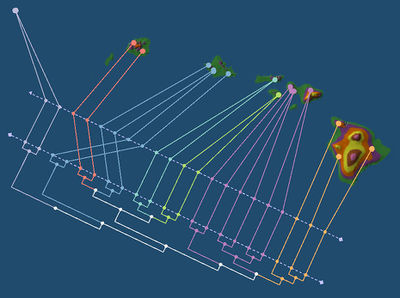
[[Image:KaydidTutorialImage.jpg|thumb|Geophylogeny of ''Banza'' katydids from the Hawaiian Islands.|400px|right]] | [[Image:KaydidTutorialImage.jpg|thumb|Geophylogeny of ''Banza'' katydids from the Hawaiian Islands.|400px|right]] | ||
| − | In this tutorial, we consider the phylogenetic tree of ''Banza'' katydids (acoustic insects) from the Hawaiian Islands recently recovered by Shapiro et al. [ | + | In this tutorial, we consider the phylogenetic tree of ''Banza'' katydids (acoustic insects) from the Hawaiian Islands recently recovered by Shapiro ''et al.'', 2006. This tutorial gives an overview of the GenGIS interface, demonstrates how to load data, and illustrates how to change the visual properties of a map, location markers, and trees. The tutorial concludes with an analysis which suggests geography has had a substantial influence on the evolution of ''Banza'' katydids. |
| + | |||
| + | * [[Katydid Tutorial | ''Banza'' Katydid tutorial]] | ||
| + | * [[Media:GenGIS_Katydid_Tutorial.mp4|Stream video tutorial]] | ||
| + | * [[Media:GenGIS_Katydids_Data.zip|Download ''Banza'' Katydid data set]] | ||
| + | |||
| + | === Tutorial 2: Marine Microbial Communities from the Atlantic Seaboard === | ||
| + | |||
| + | Here we examine samples collected as part of the Global Ocean Sampling expedition (Rusch ''et al.'', 2007) in order to investigate the influence of environmental factors on the composition of microbial communities from marine ecosystems. This tutorial demonstrates how to load data, illustrates how to change visual properties, presents a detail analysis of the geographic structure of these microbial communities, and examines the distribution of taxa within these samples. | ||
| + | |||
| + | * [[GOS Tutorial | GOS tutorial]] | ||
| + | * [[Media:GenGIS_GOS_Tutorial.mp4|Stream video tutorial]] | ||
| + | * [[Media:GenGIS_GOS_Data.zip|Download GOS data set]] | ||
| − | + | === Tutorial 3: Microbial diversity at 28 human body sites === | |
| − | + | Costello ''et al.'' (2009) collected microbial marker gene samples from 28 distinct human body sites to assess variation across sites, broad cateogories of sites (for instance, skin vs. gut) and through time. The following tutorial shows how to use plugins to perform statistical tests and generate visualizations, and how roll your own analysis scripts using Rpy2. | |
| − | * [[Media: | + | * [[Body Location Tutorial | Body Location tutorial]] |
| + | * [[Media:GenGIS_Costello_Data.zip|Download body site data set and analysis script]] | ||
| + | === Tutorial 4: The ''Ensatina eschscholtzii'' ring species === | ||
| − | + | The ''Ensatina eschscholtzii'' salamander from the western United States is a classic example of allopatric speciation. In this tutorial, we demonstrate how GenGIS can be used to analyze non-linear geographic gradients (Parks and Beiko, 2009). | |
| − | + | * [[West Coast Salamander Tutorial | Salamander tutorial]] | |
| + | * [[Media:Salamander.mp4|Stream video tutorial]] | ||
| + | * [[Media:GenGIS_Salamanders.zip|Download salamander data set]] | ||
| − | + | === Tutorial 5: Plugins === | |
| − | + | The functionality of GenGIS can be extended by writing a Python plugins. GenGIS comes with a number of Python plugins which can be accessed through the ''Plugins'' menu. Below is information about each of these plugins and instruction for developing your own plugin. | |
| + | * [[Description of GenGIS plugins | Description of GenGIS plugins]] | ||
| + | * [[Creating a GenGIS plugin | Creating a GenGIS plugin]] | ||
| + | * [[RCA_Tutorial| Tutorial showing the use of the RCA plugin]] | ||
| − | === Tutorial | + | === Tutorial 6: Custom Data Visualizations and Movies === |
We have put together a number of examples illustrating how custom data visualizations and movies can be created using the API exposed to the Python console: | We have put together a number of examples illustrating how custom data visualizations and movies can be created using the API exposed to the Python console: | ||
| − | |||
| − | |||
# An example showing how GenGIS can be used to visualize the mobility rates of HIV-1 subtype B in Europe as proposed by [http://www.retrovirology.com/content/6/1/49 Paraskevis et al. (2009)] has been posted [[HIV-1 subtype B mobility in Europe | here]]. This example includes a Python class demonstrating how a custom visualization can be made. | # An example showing how GenGIS can be used to visualize the mobility rates of HIV-1 subtype B in Europe as proposed by [http://www.retrovirology.com/content/6/1/49 Paraskevis et al. (2009)] has been posted [[HIV-1 subtype B mobility in Europe | here]]. This example includes a Python class demonstrating how a custom visualization can be made. | ||
| − | # | + | # Short examples of using the Python API to access data are available in the [[The GenGIS Manual#The Python console and API functions | GenGIS manual]]. |
| − | # | + | # Details on creating fly-through movies with GenGIS are available in the [[The GenGIS Manual#Creating fly-through movies | GenGIS manual]]. |
| − | # A | + | |
| + | === Tutorial 7: Cartogram Tutorial with ''Aneides lugubris'' === | ||
| + | |||
| + | We have put together a tutorial illustrating how to create cartograms using the Aneides lugubris salamander, which is endemic to the American West Coast. | ||
| + | # A tutorial with pictures and walkthrough | ||
| + | *[[Aneides_lugubris_Tutorial | Creating a Cartogram with GenGIS]] | ||
=== References === | === References === | ||
| − | [ | + | Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. ''Science'', 326: 1694-1697. ([http://www.sciencemag.org/content/326/5960/1694.short Abstract]) |
| + | |||
| + | Parks DH and Beiko RG. 2009. Quantitative visualizations of hierarchically organized data in a geographic context. ''Geoinformatics 2009'', Fairfax, VA. [http://ieeexplore.ieee.org/xpl/freeabs_all.jsp?reload=true&isnumber=5292806&arnumber=5293552&count=243&index=6 IEEE Xplore] | ||
| + | |||
| + | Rusch DB, Halpern AL, Sutton G, ''et al.'' .2007. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. ''PLoS Biol.'' '''5''':e77. [http://www.ncbi.nlm.nih.gov/pubmed/17355176 PubMed] | ||
| − | + | Shapiro LH, Strazanac JS, and Roderick GK. 2006 Molecular phylogeny of Banza (Orthoptera: Tettigoniidae), the endemic katydids of the Hawaiian Archipelago. ''Mol. Phylogenet. Evol.'' '''41''':53-63. [http://www.ncbi.nlm.nih.gov/pubmed/16781170 Pubmed] | |
Latest revision as of 13:37, 16 May 2017
We have put together a few tutorials to get you quickly started with GenGIS.
Contents
- 1 Short video tutorials
- 2 Walkthrough tutorials with files
- 2.1 Tutorial 0: Obtaining Custom Digital Elevation Maps (DEM)
- 2.2 Tutorial 1: Banza Katydids of the Hawaiian Islands
- 2.3 Tutorial 2: Marine Microbial Communities from the Atlantic Seaboard
- 2.4 Tutorial 3: Microbial diversity at 28 human body sites
- 2.5 Tutorial 4: The Ensatina eschscholtzii ring species
- 2.6 Tutorial 5: Plugins
- 2.7 Tutorial 6: Custom Data Visualizations and Movies
- 2.8 Tutorial 7: Cartogram Tutorial with Aneides lugubris
- 2.9 References
Short video tutorials
These videos show how different tasks can be performed in GenGIS.
Creating a basemap
Here is a short illustration of basemap creation using both raster and vector data. View Tutorial (3:25)
Querying GBIF
From version 2.1.1 you can add data directly from the Global Biodiversity Information Facility (GBIF). Here is a tutorial showing the retrieval of cougar and jaguar records from GBIF. (11:12)
Walkthrough tutorials with files
Tutorial 0: Obtaining Custom Digital Elevation Maps (DEM)
The simple way to obtain DEMs suitable for use in GenGIS is described here. An older set of instructions on how to manually contruct a custom DEM is available here. You can also obtain 2D maps compatible with GenGIS using MapMaker.
Tutorial 1: Banza Katydids of the Hawaiian Islands
In this tutorial, we consider the phylogenetic tree of Banza katydids (acoustic insects) from the Hawaiian Islands recently recovered by Shapiro et al., 2006. This tutorial gives an overview of the GenGIS interface, demonstrates how to load data, and illustrates how to change the visual properties of a map, location markers, and trees. The tutorial concludes with an analysis which suggests geography has had a substantial influence on the evolution of Banza katydids.
Tutorial 2: Marine Microbial Communities from the Atlantic Seaboard
Here we examine samples collected as part of the Global Ocean Sampling expedition (Rusch et al., 2007) in order to investigate the influence of environmental factors on the composition of microbial communities from marine ecosystems. This tutorial demonstrates how to load data, illustrates how to change visual properties, presents a detail analysis of the geographic structure of these microbial communities, and examines the distribution of taxa within these samples.
Tutorial 3: Microbial diversity at 28 human body sites
Costello et al. (2009) collected microbial marker gene samples from 28 distinct human body sites to assess variation across sites, broad cateogories of sites (for instance, skin vs. gut) and through time. The following tutorial shows how to use plugins to perform statistical tests and generate visualizations, and how roll your own analysis scripts using Rpy2.
Tutorial 4: The Ensatina eschscholtzii ring species
The Ensatina eschscholtzii salamander from the western United States is a classic example of allopatric speciation. In this tutorial, we demonstrate how GenGIS can be used to analyze non-linear geographic gradients (Parks and Beiko, 2009).
Tutorial 5: Plugins
The functionality of GenGIS can be extended by writing a Python plugins. GenGIS comes with a number of Python plugins which can be accessed through the Plugins menu. Below is information about each of these plugins and instruction for developing your own plugin.
Tutorial 6: Custom Data Visualizations and Movies
We have put together a number of examples illustrating how custom data visualizations and movies can be created using the API exposed to the Python console:
- An example showing how GenGIS can be used to visualize the mobility rates of HIV-1 subtype B in Europe as proposed by Paraskevis et al. (2009) has been posted here. This example includes a Python class demonstrating how a custom visualization can be made.
- Short examples of using the Python API to access data are available in the GenGIS manual.
- Details on creating fly-through movies with GenGIS are available in the GenGIS manual.
Tutorial 7: Cartogram Tutorial with Aneides lugubris
We have put together a tutorial illustrating how to create cartograms using the Aneides lugubris salamander, which is endemic to the American West Coast.
- A tutorial with pictures and walkthrough
References
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science, 326: 1694-1697. (Abstract)
Parks DH and Beiko RG. 2009. Quantitative visualizations of hierarchically organized data in a geographic context. Geoinformatics 2009, Fairfax, VA. IEEE Xplore
Rusch DB, Halpern AL, Sutton G, et al. .2007. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77. PubMed
Shapiro LH, Strazanac JS, and Roderick GK. 2006 Molecular phylogeny of Banza (Orthoptera: Tettigoniidae), the endemic katydids of the Hawaiian Archipelago. Mol. Phylogenet. Evol. 41:53-63. Pubmed